PROSTATE-MRI-US-BIOPSY
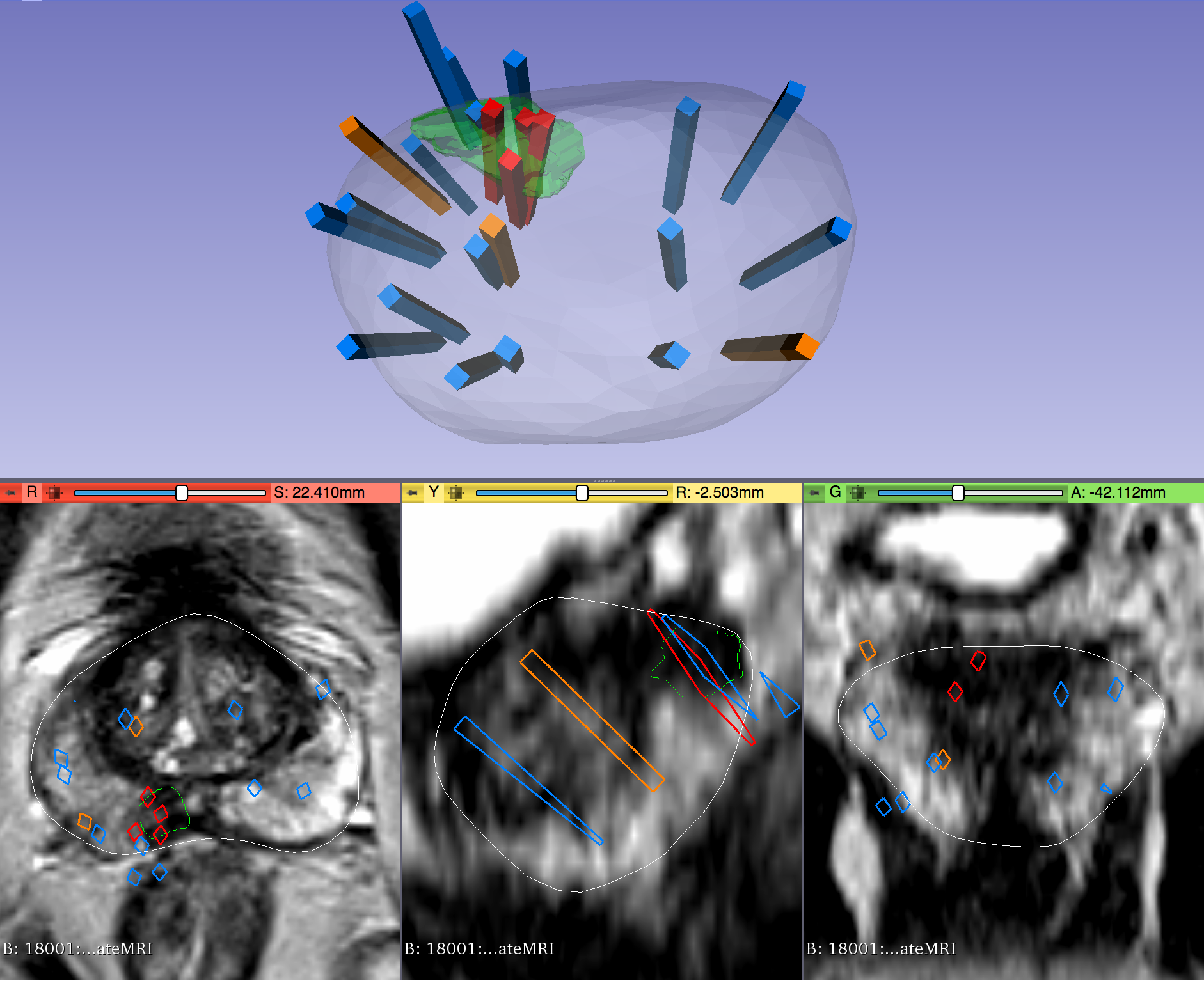
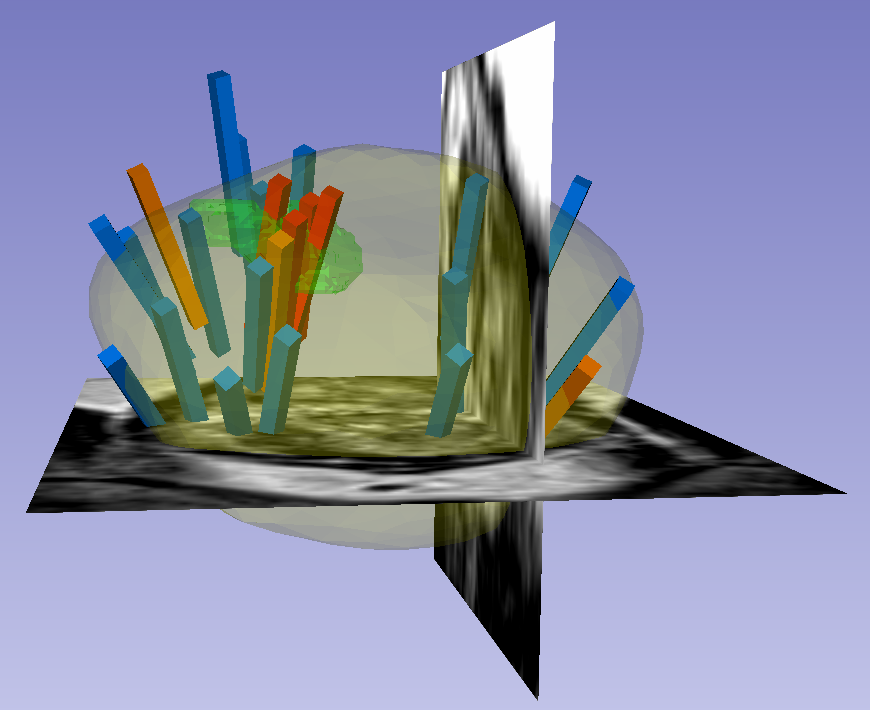
This dataset was derived from tracked biopsy sessions using the Artemis biopsy system, many of which included image fusion with MRI targets. Patients received...
[…] 3D T2:TSE with TR/TE 3800– 5040/101, and a small minority were imported from other institutions (various T2 protocols.) Ultrasound scans were performed with Hitachi Hi-Vision 5500 7.5 MHz or the Noblus C41V 2-10 MHz end-fire probe. 3D scans were acquired by rotation of the end-fire probe 200 degrees about its axis, and interpolating to resample the volume with isotropic resolution. Patients with suspicion of prostate cancer due to elevated PSA and/or suspicious imaging findings were consecutively accrued. Any consented patient who underwent or had planned to receive a routine, standard-of-care prostate biopsy at the UCLA Clark Urology Center was included. Note: Some Private Tags in this collection are critical to properly displaying the STL surface and the Prostate anatomy. Private Tag (1129,”Eigen Artemis”,1016) DS VoxelSize is especially important for multi-frame US cases. Acknowledgements We would like to acknowledge the individuals and institutions that have provided data for this collection: This work was supported in part by the National Cancer Institute Award R01CA158627, Prostate Cancer SPORE at University of California-Los Angeles P50CA092131, the Beckman Coulter Foundation, Jean Perkins Foundation, and Steven C. Gordon Family Foundation Harmonization of the components of this dataset, including into standard DICOM representation, was supported in part by the NCI Imaging Data Commons consortium. NCI Imaging Data Commons consortium is supported by the contract number 19X037Q from Leidos Biomedical Research under Task Order HHSN26100071 from NCI Data Access Detailed Description Citations & Data Usage Policy Versions Data Access Data Type Download all or Query/Filter License Images (DICOM) 79.4 (GB) Download Search (Download requires the NBIA Data Retriever) CC BY 4.0 Target Data (XLSX, 131 KB) Download CC BY 4.0 Biopsy Data (XLSX, 4.25 MB) Download CC BY 4.0 STL Files (ZIP, 274 MB) Download (Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) CC BY 4.0 Biopsy Overlays (ZIP, 333 MB) Download (Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) CC BY 4.0 Click the Versions tab for more info about data releases. Additional Resources for this Dataset The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data. Imaging Data Commons (IDC) (Imaging Data) Detailed Description Image Statistics Radiology Image Statistics Modalities MR, US Number of Participants 1,151 Number of Studies 2,799 Number of Series 4,819 Number of Images 102,397 Images Size (GB) 79.4 Instructions for Proper Use of the STL Files The 3D surfaces of this dataset are in stereolithography format (binary representation). They are segmentations representing anatomic regions within the corresponding ultrasound (US) or MRI images. Each file is named “Prostate-MRI-US-Biopsy-XXXX-SURFACETYPE-seriesUID-YYYY.STL” where “XXXX” is the anonymized patient number and “YYYY” is the series instance UID of corresponding DICOM images. “SURFACETYPE” can be one of two identifiers, summarized below. If “SURFACETYPE” is “ProstateSurface,” then the STL file is a segmentation of the prostate gland. These were contoured semi-automatically with Profuse (for MRI) or the Artemis fusion biopsy system (for US), both products of Eigen (Grass Valley, CA). Using these tools, the user manually traced the prostate on one or more image planes, and the remainder of gland was automatically segmented in 3D. The user was then able to adjust vertices of the 3D segmentation as necessary. These segmentations are generally of good quality, though their accuracy is user-dependent. If “SURFACETYPE” is “Target,” then the STL file is a segmentation of a Region of Interest (ROI) suspicious for harboring prostate cancer. Zero, one, or more than one ROI may be present, and thus are designated “Target1,” “Target2,” etc. The ROIs corresponding to MRI were traced manually by a radiologist based on their interpretation of the patient’s multiparametric MRI study. The ROIs corresponding to US images were acquired by registering the MRI prostate surface nonrigidly and semi-automatically to the US prostate surface. Therefore, the US ROIs are subject to misregistration, with average accuracy of 3-4 mm. Note that proximity to an ROI increases the likelihood of, but does not guarantee, prostate cancer. Also note that radiologists assigned each ROI a Likert-like suspicion score of 1-5, with 1 corresponding to very low suspicion of prostate cancer and 5 corresponding to very high suspicion of prostate cancer. These scores, which are roughly equivalent to the “PIRADS Version 2” scoring system, can be found in a spreadsheet included with the dataset. The STL files can be visualized in 3D Slicer as follows: Import the surface(s) either by drag-and-dropping or with File->Add Data Within the “Models” module, adjust Display->Opacity of the prostate surface in order to visualize any ROI(s) within To visualize surfaces in the context of corresponding MRI or US images, load the corresponding image series and then toggle the Slice Display->Visible radio button within the “models” module To visualize surfaces in the context of biopsy cores, import the corresponding 3D slicer file named “BiopsyVectors.mrml” by drag-and-dropping or with File->Add Data. The STL files, which are written in a standard format, can also be imported and visualized with most computer-aided design (CAD) and 3D printing software. Note that, if quantitative analysis is desired, the following transform was applied to US surfaces for 3D slicer compatibility and may need to be reversed for visualization alongside MRI or US images using other software. Transform (which has been applied to all ultrasound data): Inverse Transform (use to revert ultrasound data to original coordinates): Instructions for Proper Use of the “Biopsy Overlay” Files Biopsy vectors, prostate surfaces, and regions of interest can be imported to 3D Slicer in a single step using the collection’s “Biopsy Overlay” files. Overlay files associated with ultrasound images are organized into folders with the naming convention “Prostate-MRI-US-Biopsy-XXXX-BXus-seriesUID-####” where “XXXX” is the anonymized patient number and “####” is the series instance UID of corresponding DICOM images. Files associated with MR images have an analogous naming convention “Prostate-MRI-US-Biopsy-XXXX-BXmr-seriesUID-####”, but are appended with “(US-date-YYYYMMDD)” where ‘YYYYMMDD’ is the de-identified acquisition date of the ultrasound series registered to MRI during biopsy. This is specified because in some cases multiple ultrasound-guided biopsies were registered to the same MRI series. Within each folder of “Biopsy Overlays” is a file named “BiopsyVectors.mrml”. This can be imported directly into 3D Slicer by either by drag-and-dropping or with File->Add Data. The prostate surface, any ROIs present, and all biopsy vectors will then be displayed within 3D Slicer. The biopsy cores, represented as “line” fiducials, are color-coded as BLUE = benign, ORANGE = Gleason Grade 3+3, and RED = Gleason Grade ≥ 3+4. The prostate is shown as WHITE, and ROIs are GREEN. To visualize these overlays in the context of corresponding MRI or US images, load the corresponding image series to 3D Slicer using File->DICOM. Instructions for Proper Use […]