NSCLC Radiogenomics | NSCLC Radiogenomics
DOI: 10.7937/K9/TCIA.2017.7hs46erv | Data Citation Required | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Chest | Human | 211 | SEG, CT, PT | Non-small Cell Lung Cancer | Clinical, Image Analyses, Genomics | Public, Complete | 2021/06/01 |
Summary
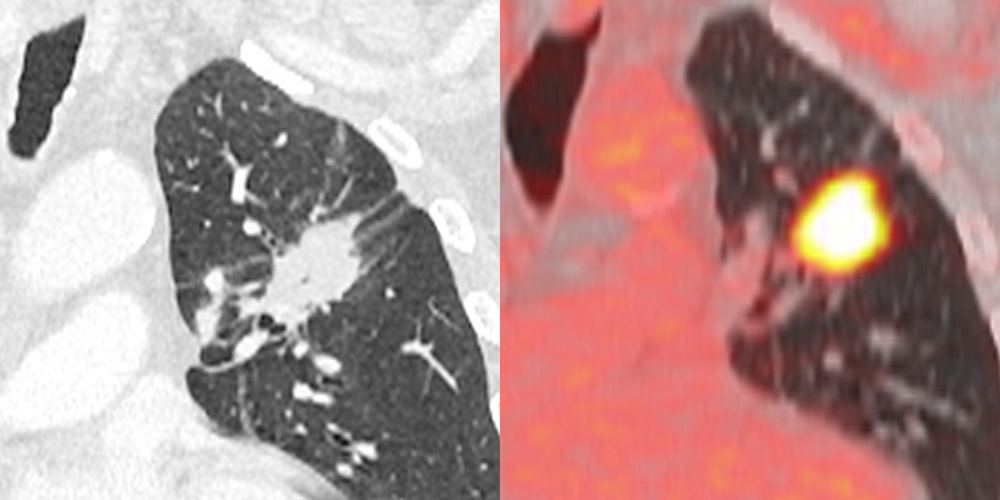
Medical image biomarkers of cancer promise improvements in patient care through advances in precision medicine. Compared to genomic biomarkers, image biomarkers provide the advantages of being a non-invasive procedure, and characterizing a heterogeneous tumor in its entirety, as opposed to limited tissue available for biopsy. We developed a unique radiogenomic dataset from a Non-Small Cell Lung Cancer (NSCLC) cohort of 211 subjects. The dataset comprises Computed Tomography (CT), Positron Emission Tomography (PET)/CT images, semantic annotations of the tumors as observed on the medical images using a controlled vocabulary, segmentation maps of tumors in the CT scans, and quantitative values obtained from the PET/CT scans. Imaging data are also paired with gene mutation, RNA sequencing data from samples of surgically excised tumor tissue, and clinical data, including survival outcomes. This dataset was created to facilitate the discovery of the underlying relationship between genomic and medical image features, as well as the development and evaluation of prognostic medical image biomarkers. Further details regarding this data-set may be found in Bakr, et. al, Sci Data. 2018 Oct 16;5:180202. doi: 10.1038/sdata.2018.202, https://www.ncbi.nlm.nih.gov/pubmed/30325352. For scientific and other inquiries about this dataset, please contact TCIA's Helpdesk.
Data Access
Version 4: Updated 2021/06/01
- Added missing image studies for the following cases: R01-009 (CT), R01-100 (PET/CT), and R01-111 (PET/CT).
- SUV conversion factor DICOM tag (7053,1000) was added for the following Philips PET images: R01-074, R01-077, R01-079, R01-089, R01-98 and R01-137.
| Title | Data Type | Format | Access Points | Subjects | License | |||
|---|---|---|---|---|---|---|---|---|
| Images and Segmentations | SEG, CT, PT | DICOM | Download requires NBIA Data Retriever |
211 | 395 | 1,351 | 286,754 | CC BY 3.0 |
| AIM Annotations | XML and ZIP | CC BY 3.0 | ||||||
| Clinical Data | CSV | CC BY 3.0 |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
- IDC Zenodo community dataset: Image segmentations produced by BAMF under the AIMI Annotations initiative
The following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- RNA sequence data (Note: 130 subject subset)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Bakr, S., Gevaert, O., Echegaray, S., Ayers, K., Zhou, M., Shafiq, M., Zheng, H., Zhang, W., Leung, A., Kadoch, M., Shrager, J., Quon, A., Rubin, D., Plevritis, S., & Napel, S. (2017). Data for NSCLC Radiogenomics (Version 4) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2017.7hs46erv |
Detailed Description
This collection was originally submitted to TCIA as a 26 subject pilot data set. You can learn more about that subset of the collection in the following Analysis Results publication:
Data Citation
Napel, Sandy, & Plevritis, Sylvia K. (2014). NSCLC Radiogenomics: Initial Stanford Study of 26 Cases. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2014.X7ONY6B1
Related Publications
Publication Citation |
|
|
Bakr, S., Gevaert, O., Echegaray, S., Ayers, K., Zhou, M., Shafiq, M., Zheng, H., Benson, J. A., Zhang, W., Leung, A., Kadoch, M., Hoang, C. D., Shrager, J., Quon, A., Rubin, D. L., Plevritis, S. K., & Napel, S. (2018). A radiogenomic dataset of non-small cell lung cancer. Scientific data, 5, 180202. https://doi.org/10.1038/sdata.2018.202 |
Publication Citation |
|
|
Gevaert, O., Xu, J., Hoang, C. D., Leung, A. N., Xu, Y., Quon, A., … Plevritis, S. K. (2012, August). Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology. Radiological Society of North America (RSNA). http://doi.org/10.1148/radiol.12111607 |
Research Community Publications
TCIA maintains a list of publications that leverage TCIA data.
- Aonpong, P., Iwamoto, Y., Han, X. H., Lin, L., & Chen, Y. W. (2021). Improved Genotype-Guided Deep Radiomics Signatures for Recurrence Prediction of Non-Small Cell Lung Cancer Annu Int Conf IEEE Eng Med Biol Soc, 2021, 3561-3564. doi:https://doi.org/10.1109/EMBC46164.2021.9630703
- Aonpong, P., Iwamoto, Y., Wang, W., Lin, L., & Chen, Y.-W. (2020). Hand-Crafted and Deep Learning-Based Radiomics Models for Recurrence Prediction of Non-Small Cells Lung Cancers. Innovation in Medicine and Healthcare, 192, 135-144. doi:https://doi.org/10.1007/978-981-15-5852-8_13
- Aonpong, P., Iwamoto, Y., Wang, W., Lin, L., & Chen, Y.-W. (2021). Genomics-Based Models for Recurrence Prediction of Non-small Cells Lung Cancers. Paper presented at the KES International Conferences on Innovation in Medicine and Healthcare (KES-InMed-21)
- Brummer, A. B., & Savage, V. M. (2021). Cancer as a Model System for Testing Metabolic Scaling Theory. Frontiers in Ecology and Evolution, 9. doi:10.3389/fevo.2021.691830
- Caruso, C. M., Guarrasi, V., Cordelli, E., Sicilia, R., Gentile, S., Messina, L., . . . Soda, P. (2022). A Multimodal Ensemble Driven by Multiobjective Optimisation to Predict Overall Survival in Non-Small-Cell Lung Cancer. J Imaging, 8(11). doi:https://doi.org/10.3390/jimaging8110298
- Chen, W., Qiao, X., Yin, S., Zhang, X., & Xu, X. (2022). Integrating Radiomics with Genomics for Non-Small Cell Lung Cancer Survival Analysis. J Oncol, 2022, 5131170. doi:https://doi.org/10.1155/2022/5131170
- Cho, H.-h., Lee, H. Y., Kim, E., Lee, G., Kim, J., Kwon, J., & Park, H. (2021). Radiomics-guided deep neural networks stratify lung adenocarcinoma prognosis from CT scans. Communications Biology, 4(1), 1-12. doi:10.1038/s42003-021-02814-7
- Choi, J., Cho, H. H., Kwon, J., Lee, H. Y., & Park, H. (2021). A Cascaded Neural Network for Staging in Non-Small Cell Lung Cancer Using Pre-Treatment CT. Diagnostics (Basel), 11(6). doi:10.3390/diagnostics11061047
- Christie, J. R., Daher, O., Abdelrazek, M., Romine, P. E., Malthaner, R. A., Qiabi, M., . . . Mattonen, S. A. (2022). Predicting recurrence risks in lung cancer patients using multimodal radiomics and random survival forests. J Med Imaging (Bellingham), 9(6), 066001. doi:https://doi.org/10.1117/1.JMI.9.6.066001
- Gui, D., Song, Q., Song, B., Li, H., Wang, M., Min, X., & Li, A. (2022). AIR-Net: A novel multi-task learning method with auxiliary image reconstruction for predicting EGFR mutation status on CT images of NSCLC patients. Comput Biol Med, 141, 105157. doi:10.1016/j.compbiomed.2021.105157
- Hosny, A., Bitterman, D. S., Guthier, C. V., Qian, J. M., Roberts, H., Perni, S., . . . Mak, R. H. (2022). Clinical validation of deep learning algorithms for radiotherapy targeting of non-small-cell lung cancer: an observational study. The Lancet Digital Health, 4(9), e657-e666. doi:10.1016/S2589-7500(22)00129-7
- Ju, H. M., Kim, B. C., Lim, I., Byun, B. H., & Woo, S. K. (2023). Estimation of an Image Biomarker for Distant Recurrence Prediction in NSCLC Using Proliferation-Related Genes. Int J Mol Sci, 24(3). doi:https://doi.org/10.3390/ijms24032794
- Kadoya, N., Tanaka, S., Kajikawa, T., Tanabe, S., Abe, K., Nakajima, Y., . . . Jingu, K. (2020). Homology-based radiomic features for prediction of the prognosis of lung cancer based on CT-based radiomics. Med Phys. doi:10.1002/mp.14104
- Koyasu, S., Nishio, M., Isoda, H., Nakamoto, Y., & Togashi, K. (2020). Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on (18)F FDG-PET/CT. Ann Nucl Med, 34(1), 49-57. doi:https://doi.org/10.1007/s12149-019-01414-0
- Lee, S. H., Cho, H. H., Kwon, J., Lee, H. Y., & Park, H. (2021). Are radiomics features universally applicable to different organs? Cancer Imaging, 21(1), 31. doi:10.1186/s40644-021-00400-y
- Li, J., Qiu, Z., Zhang, C., Chen, S., Wang, M., Meng, Q., . . . Zhang, X. (2022). ITHscore: comprehensive quantification of intra-tumor heterogeneity in NSCLC by multi-scale radiomic features. Eur Radiol. doi:10.1007/s00330-022-09055-0
- Lin, P., Lin, Y. Q., Gao, R. Z., Wan, W. J., He, Y., & Yang, H. (2023). Integrative radiomics and transcriptomics analyses reveal subtype characterization of non-small cell lung cancer. Eur Radiol. doi:https://doi.org/10.1007/s00330-023-09503-5
- Mattonen, S. A., Davidzon, G. A., Benson, J., Leung, A. N. C., Vasanawala, M., Horng, G., . . . Nair, V. S. (2019). Bone Marrow and Tumor Radiomics at (18)F-FDG PET/CT: Impact on Outcome Prediction in Non-Small Cell Lung Cancer. Radiology, 190357. doi:10.1148/radiol.2019190357
- Mienye, I. D. (2021). Improved Machine Learning Algorithms with Application to Medical Diagnosis. (Dissertation). University of Johannesburg,
- Mienye, I. D., Sun, Y., & Wang, Z. (2020). Improved Predictive Sparse Decomposition Method with Densenet for Prediction of Lung Cancer. International Journal of Computing, 19(4), 533-541. doi:10.47839/ijc.19.4.1986
- Moitra, D., & Kr. Mandal, R. (2020). Classification of non-small cell lung cancer using one-dimensional convolutional neural network. Expert Systems with Applications, 159, 113564. doi:https://doi.org/10.1016/j.eswa.2020.113564
- Moitra, D., & Mandal, R. K. (2019). Automated AJCC (7th edition) staging of non-small cell lung cancer (NSCLC) using deep convolutional neural network (CNN) and recurrent neural network (RNN). Health Inf Sci Syst, 7(1), 14. doi:10.1007/s13755-019-0077-1
- Moitra, D., & Mandal, R. K. (2020). Prediction of Non-small Cell Lung Cancer Histology by a Deep Ensemble of Convolutional and Bidirectional Recurrent Neural Network. Journal of Digital Imaging, 1-8. doi:10.1007/s10278-020-00337-x
- Moitra, D., & Mandal, R. K. (2022). Classification of malignant tumors by a non-sequential recurrent ensemble of deep neural network model. Multimed Tools Appl, 1-19. doi:10.1007/s11042-022-12229-z
- Morgado, J., Pereira, T., Silva, F., Freitas, C., Negrão, E., de Lima, B. F., . . . Oliveira, H. P. (2021). Machine Learning and Feature Selection Methods for EGFR Mutation Status Prediction in Lung Cancer. Applied Sciences, 11(7), 3273. doi:10.3390/app11073273
- Mukherjee, P., Zhou, M., Lee, E., Schicht, A., Balagurunathan, Y., Napel, S., . . . Gevaert, O. (2020). A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nature Machine Intelligence, 2, 274-282. doi:https://doi.org/10.1038/s42256-020-0173-6
- Ninomiya, K., Arimura, H., Chan, W. Y., Tanaka, K., Mizuno, S., Muhammad Gowdh, N. F., . . . Ng, K. H. (2021). Robust radiogenomics approach to the identification of EGFR mutations among patients with NSCLC from three different countries using topologically invariant Betti numbers. PLoS One, 16(1), e0244354. doi:https://doi.org/10.1371/journal.pone.0244354
- Pasini, G., Stefano, A., Russo, G., Comelli, A., Marinozzi, F., & Bini, F. (2023). Phenotyping the Histopathological Subtypes of Non-Small-Cell Lung Carcinoma: How Beneficial Is Radiomics? Diagnostics (Basel), 13(6). doi:https://doi.org/10.3390/diagnostics13061167
- Primakov, S. P., Ibrahim, A., van Timmeren, J. E., Wu, G., Keek, S. A., Beuque, M., . . . Lambin, P. (2022). Automated detection and segmentation of non-small cell lung cancer computed tomography images. Nature Communications, 13(1), 3423. doi:10.1038/s41467-022-30841-3
- Pu, L., Gezer, N. S., Ashraf, S. F., Ocak, I., Dresser, D. E., & Dhupar, R. (2022). Automated segmentation of five different body tissues on computed tomography using deep learning. Med Phys. doi:https://doi.org/10.1002/mp.15932
- Saad, M., & Choi, T.-S. (2018). Computer-assisted subtyping and prognosis for non-small cell lung cancer patients with unresectable tumor. Computerized Medical Imaging and Graphics, 67, 1-8. doi:10.1016/j.compmedimag.2018.04.003
- Shi, F., Hu, W., Wu, J., Han, M., Wang, J., Zhang, W., . . . Shen, D. (2022). Deep learning empowered volume delineation of whole-body organs-at-risk for accelerated radiotherapy. Nat Commun, 13(1), 6566. doi:https://doi.org/10.1038/s41467-022-34257-x
- Shiri, I., Amini, M., Nazari, M., Hajianfar, G., Haddadi Avval, A., Abdollahi, H., . . . Zaidi, H. (2022). Impact of feature harmonization on radiogenomics analysis: Prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput Biol Med, 142, 105230. doi:https://doi.org/10.1016/j.compbiomed.2022.105230
- Shiri, I., Maleki, H., Hajianfar, G., Abdollahi, H., Ashrafinia, S., Hatt, M., . . . Rahmim, A. (2020). Next-generation radiogenomics sequencing for prediction of EGFR and KRAS mutation status in NSCLC patients using multimodal imaging and machine learning algorithms. Molecular Imaging and Biology, 22(4), 1132-1148. doi:10.1007/s11307-020-01487-8
- Shiri, I., Vafaei Sadr, A., Akhavan, A., Salimi, Y., Sanaat, A., Amini, M., . . . Zaidi, H. (2022). Decentralized collaborative multi-institutional PET attenuation and scatter correction using federated deep learning. Eur J Nucl Med Mol Imaging, 1-17. doi:https://doi.org/10.1007/s00259-022-06053-8
- Thomas, R., Schalck, E., Fourure, D., Bonnefoy, A., & Cervera-Marzal, I. (2021). 2Be3-Net: Combining 2D and 3D Convolutional Neural Networks for 3D PET Scans Predictions. Paper presented at the International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2021).
- Torres, F. S., Akbar, S., Raman, S., Yasufuku, K., Schmidt, C., Hosny, A., . . . Leighl, N. B. (2021). End-to-End Non-Small-Cell Lung Cancer Prognostication Using Deep Learning Applied to Pretreatment Computed Tomography. JCO Clin Cancer Inform, 5, 1141-1150. doi:10.1200/cci.21.00096
- Trebeschi, S., Bodalal, Z., van Dijk, N., Boellaard, T. N., Apfaltrer, P., Tareco Bucho, T. M., . . . Beets-Tan, R. G. H. (2021). Development of a Prognostic AI-Monitor for Metastatic Urothelial Cancer Patients Receiving Immunotherapy. Front Oncol, 11, 637804. doi:10.3389/fonc.2021.637804
- Tripathi, S., Moyer, E. J., Augustin, A. I., Zavalny, A., Dheer, S., Sukumaran, R., . . . Kim, E. (2022). RadGenNets: Deep learning-based radiogenomics model for gene mutation prediction in lung cancer. Informatics in Medicine Unlocked, 33. doi:https://doi.org/10.1016/j.imu.2022.101062
- Wang, S. (2022). Multi-Modality Automatic Lung Tumor Segmentation Method Using Deep Learning and Radiomics. (Ph.D. Dissertation). Virginia Commonwealth University, Virginia, USA.
- Wang, Z., Yang, C., Han, W., Sui, X., Zheng, F., Xue, F., . . . Jiang, J. (2022). Quantifying lung cancer heterogeneity using novel CT features: a cross-institute study. Insights Imaging, 13(1), 82. doi:10.1186/s13244-022-01204-9
- Yousefi, B., Jahani, N., LaRiviere, M. J., Cohen, E., Hsieh, M.-K., Luna, J. M., . . . Kontos, D. (2019). Correlative hierarchical clustering-based low-rank dimensionality reduction of radiomics-driven phenotype in non-small cell lung cancer. Paper presented at the SPIE Medical Imaging, San Diego, California, United States.
If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 3: Updated 2020/11/10
- A new version of RO1-023 was created to correct a cranial-caudal flip of the segmentation of the CT volume (483 images) and associated Segmentation object. The UIDs of the other scans were updated to preserve Study level consistency but were otherwise unmodified. The referenced UIDs within the AIM object for RO1-023 were updated and renamed to RO1-023v1.
- RO1-038 was updated to remove a coronal slice at the start of the of the CT volume. This created difficulty for some software to determine slice spacing.
| Title | Data Type | Format | Access Points | License | ||||
|---|---|---|---|---|---|---|---|---|
| Images | DICOM | Download requires NBIA Data Retriever |
||||||
| AIM Annotations | XML and ZIP | |||||||
| Clinical Data | CSV |
Version 2: Updated 2017/02/28
| Title | Data Type | Format | Access Points | License | ||||
|---|---|---|---|---|---|---|---|---|
| Images | DICOM | Download requires NBIA Data Retriever |
||||||
| AIM Annotations | XML and ZIP | |||||||
| Clinical Data | CSV |
Version 1: Updated 2015/12/22
This collection was originally submitted to TCIA as a 26 subject pilot data set. You can learn more about that subset of the collection in the following Analysis Results publication:
