PROSTATEx-Seg-HiRes | High Resolution Prostate Segmentations for the ProstateX-Challenge
DOI: 10.7937/tcia.2019.deg7zg1u | Data Citation Required | Analysis Result
| Location | Subjects | Size | Updated | |||
|---|---|---|---|---|---|---|
| Prostate | Prostate | 66 | Organ segmentations | 2020/09/18 |
Summary
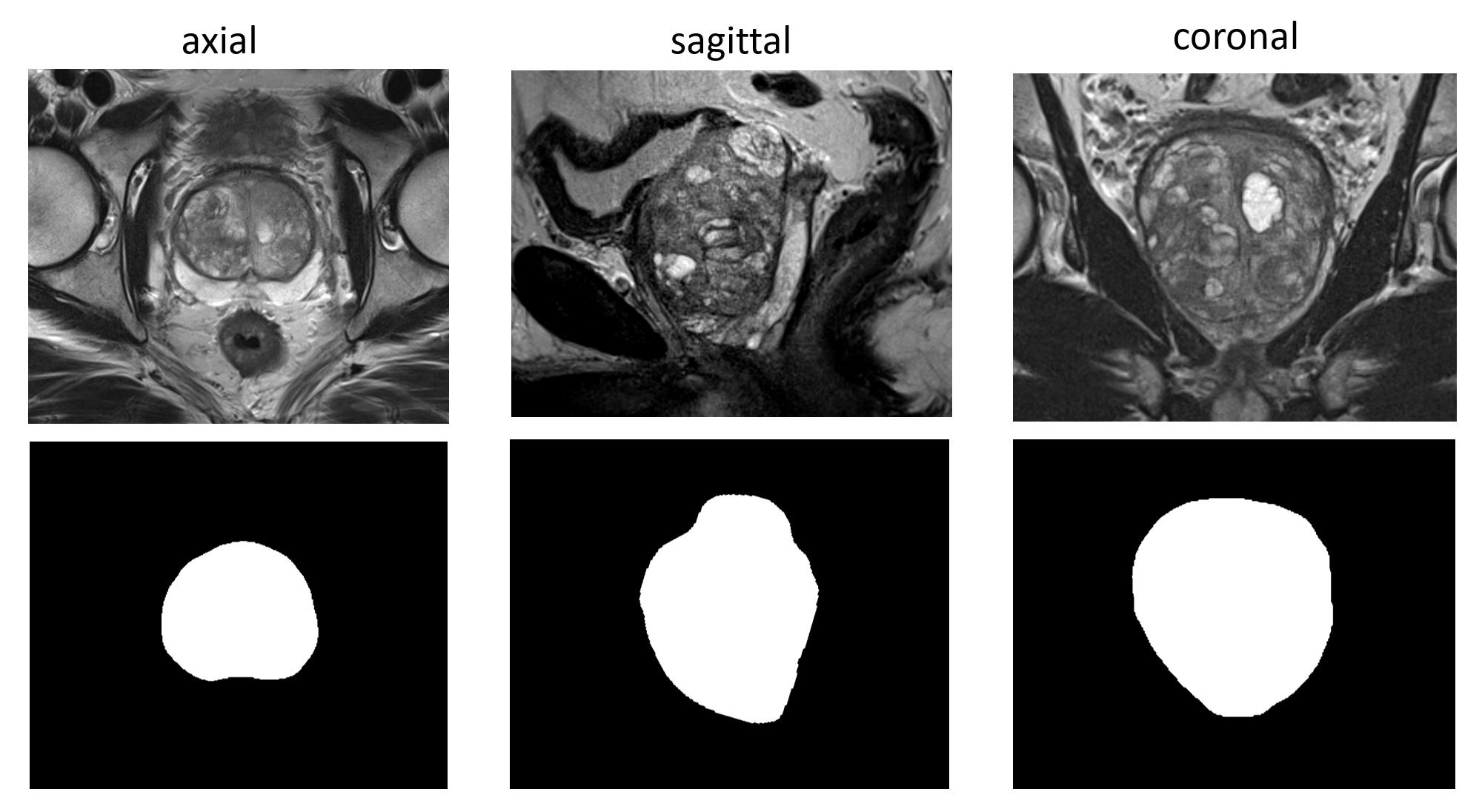
We created 66 high resolution segmentations for randomly chosen T2-weighted volumes of the SPIE-AAPM-NCI PROSTATEx Challenges (PROSTATEx). The high resolution segmentations were obtained by considering the three scan directions: for each scan direction (axial, sagittal, coronal), the gland was manually delineated by a medical student, followed by a review and corrections of an expert urologist. These three anisotropic segmentations were fused to one isotropic segmentation by means of shape-based interpolation in the following manner: (1) The signed distance transformation of the three segmentations is computed. (2) The anisotropic distance volumes are transformed into an isotropic high-resolution representation with linear interpolation. (3) By averaging the distances, smoothing and thresholding them at zero, we obtained the fused segmentation. The resulting segmentations were manually verified and corrected further by the expert urologist if necessary. Serving as ground truth for training CNNs, these segmentations have the potential to improve the segmentation accuracy of automated algorithms. By considering not only the axial scans but also sagittal and coronal scan directions, we aimed to have higher fidelity of the segmentations especially at the apex and base regions of the prostate. The segmentations to standard DICOM representation were created with dcmqi
Data Access
Version 1: Updated 2020/09/18
| Title | Data Type | Format | Access Points | Subjects | License | |||
|---|---|---|---|---|---|---|---|---|
| Segmentations | SEG | DICOM | Download requires NBIA Data Retriever |
66 | 66 | 66 | 66 | CC BY 3.0 |
Collections Used In This Analysis Result
| Title | Data Type | Format | Access Points | Subjects | License | |||
|---|---|---|---|---|---|---|---|---|
| Corresponding Original MR Images from PROSTATEx | MR | DICOM | Requires NBIA Data Retriever |
66 | 66 | 66 | 1,361 | CC BY 3.0 |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Schindele, D., Meyer, A., Von Reibnitz, D. F., Kiesswetter, V., Schostak, M., Rak, M., & Hansen, C. (2020). High Resolution Prostate Segmentations for the ProstateX-Challenge [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.2019.deg7zg1u
|
Acknowledgements
- This work has been funded by the EU and the federal state of Saxony-Anhalt, Germany under grant number ZS/2016/08/80388.
Related Publications
Publication Citation |
|
|
Meyer, A., Chlebus, G., Rak, M., Schindele, D., Schostak, M., van Ginneken, B., Schenk, A., Meine, H., Hahn, H. K., Schreiber, A., & Hansen, C. (2020). Anisotropic 3D Multi-Stream CNN for Accurate Prostate Segmentation from Multi-Planar MRI. Computer Methods and Programs in Biomedicine, 105821. https://doi.org/10.1016/j.cmpb.2020.105821 |
Research Community Publications
TCIA maintains a list of publications that leverage TCIA data. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
