Yale-Brain-Mets-Longitudinal | Yale longitudinal dataset of brain metastases on MRI with associated clinical data
DOI: 10.7937/3yat-e768 | Data Citation Required | 943 Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Brain | Human | 1,430 | MR, Demographic | Metastatic disease | Public, Complete | 2025/05/30 |
Summary
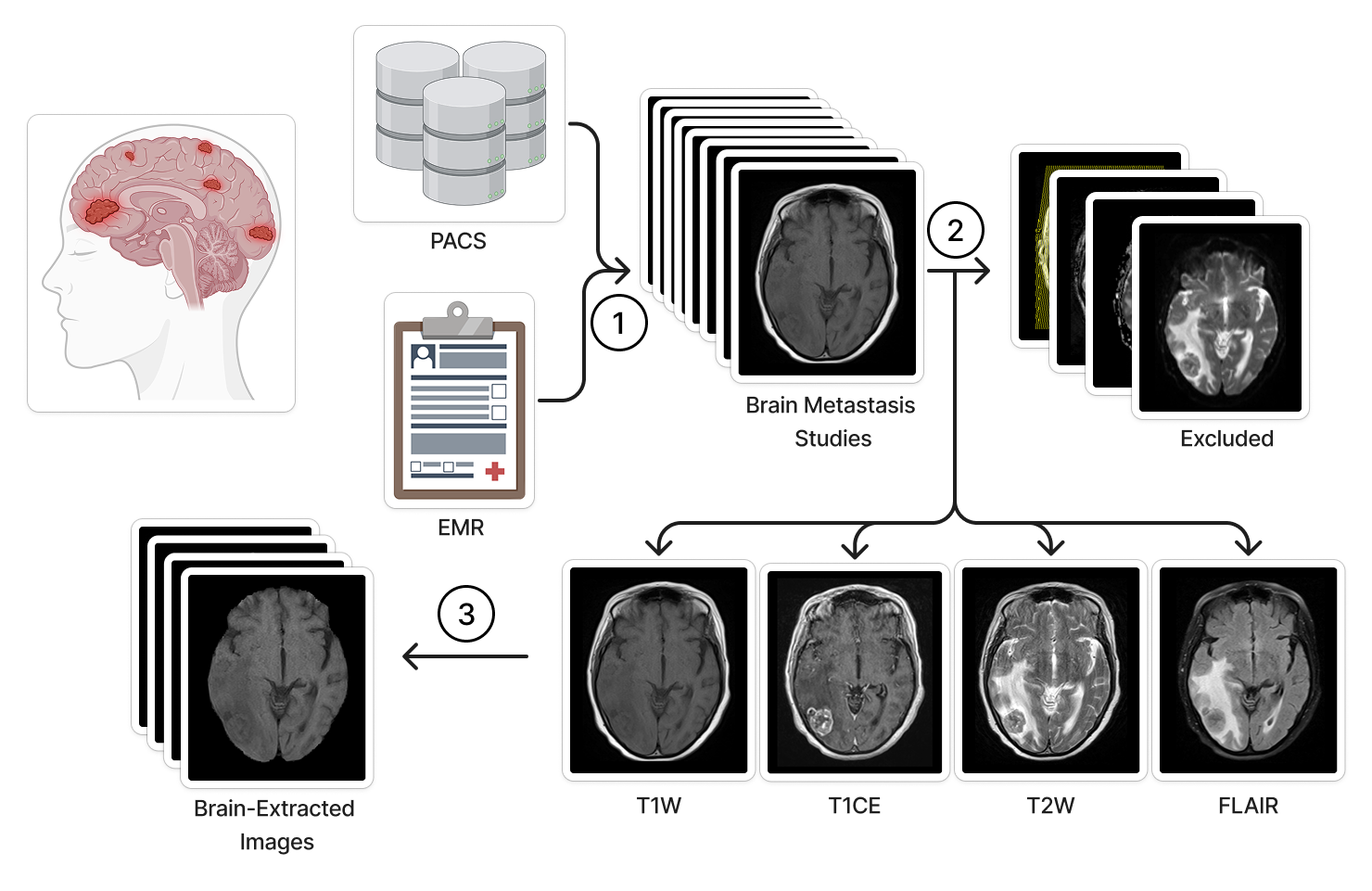
We present a dataset of 11,892 longitudinal brain MRI studies from 1,430 patients with clinically confirmed brain metastases. T1-weighted pre-contrast, T1-weighted post-contrast, T2-weighted, and fluid-attenuated inversion recovery MRI sequence images are provided in NIfTI format. Additionally, an Excel spreadsheet with patient demographic information, scanner details, and image acquisition parameters are provided. This dataset will facilitate the development of AI models to assist in the long-term management of patients with brain metastasis. Brain metastases are associated with significant morbidity, (Achrol 2019, Sacks 2020, Lamba 2021) necessitating frequent radiologic assessment in collaboration with neuro-oncologists to evaluate treatment response and disease progression. Magnetic resonance imaging (MRI) is a cornerstone in the management of central nervous system metastases, (Brenner 2022, Lin 2015, Vogelbaum 2022, Le Rhun 2021, Patil 2017, Kraft 2019, Aldawsari 2023) providing critical insights over time. (Kang 2009, Friedman 2001, Lunsford 1998) Artificial intelligence (AI) emerged as a valuable tool for prognosis and treatment planning in neuro-oncology. (Cassinelli Petersen 2022, Aneja 2019, Aboian 2022, Aboian 2022, Rudie 2021, Xue 2020) However, the creation of widely applicable clinical models is constrained by the scarcity of large-scale, heterogeneous datasets. (Aneja 2019, Rudie 2019) This underscores the need for a longitudinal imaging dataset that captures a diverse range of imaging patterns, scanner technologies, and acquisition techniques. In response to this gap, we introduce a dataset spanning nearly 20 years, including pre- and post-treatment imaging across four essential MRI sequences. To our knowledge, this is the largest publicly available MRI dataset of patients with brain metastases. By providing open access to this resource, we hope to enable diverse research applications, from conventional radiologic investigations to state-of-the-art machine learning approaches, ultimately contributing to better patient outcomes and a more comprehensive understanding of brain metastases. The inclusion of both imaging and clinical data makes this dataset a valuable asset for researchers in oncology, neuroradiology, and data science. The following subsections provide information about how the data were selected, acquired and prepared for publication, approximate date range of imaging studies. The electronic medical record (EMR) system at Yale New Haven Hospital was searched for MRI scans performed between 2004 and 2023 that evaluated brain metastases. This automated query initially retrieved 46,364 MRI studies from 7,111 patients with potential intracranial metastatic disease. A subsequent manual review of the electronic health record (EHR) excluded cases lacking radiologic or pathologic confirmation of brain metastases. To ensure consistency, only MRI exams containing axial T1-weighted (T1W), contrast-enhanced T1-weighted (T1CE), T2-weighted (T2), or fluid-attenuated inversion recovery (FLAIR) sequences were selected. For patients who underwent treatments targeting brain metastases—such as stereotactic radiosurgery, whole-brain radiotherapy, or surgical resection—pre-treatment scans taken within 30 days before treatment initiation were retained, along with all follow-up imaging to enable longitudinal analysis of disease progression and treatment effects. After these refinements, the final dataset comprised 11,892 MRI studies from 1,430 patients with confirmed brain metastases. This retrospective study was approved by the Institutional Review Board of Yale University on 10/01/2020, protocol 2000029055. Radiology: Most of the MRI scans were obtained using 1.5T or 3T scanners manufactured by Siemens Healthineers or General Electric Healthcare. Image data and associated metadata were extracted through the application programming interface of Visage (Visage 7, Visage Imaging, Inc., San Diego, CA). DICOM metadata enabled the retrieval of key imaging parameters, including study location, scanner manufacturer, scanner model, magnetic field strength, acquisition type (2D vs. 3D), sequence designation, slice thickness, slice spacing, repetition time, echo time, and inversion time. A comprehensive breakdown of these acquisition parameters for each scan is available in the accompanying Excel file. Clinical: Patient baseline information was extracted from the EMR for each study time point. The recorded data include the patient's age at the time of imaging, sex, study date. All data were retrieved as of December 2023. MRI Sequence Selection and Standardization: The MRI sequences T1W, T1CE, T2, and FLAIR were chosen for inclusion due to their essential role in evaluating brain metastases, as they provide complementary imaging characteristics critical for diagnosis and longitudinal assessment. To ensure consistency across the dataset, MRI sequence names were standardized to address variations in DICOM metadata arising from differences in scanners, radiology technicians, imaging sites, and longitudinal studies. A manual review of studies guided the development of a rules-based image classifier and validation process. Images were filtered based on factors such as orientation, acquisition technique, contrast enhancement, and spin echo variations to retain only relevant sequences. Additionally, redundant sequence identifiers were removed to streamline naming conventions. This structured approach ensured precise inclusion and uniform labeling of MRI sequences, enhancing the dataset’s reliability for longitudinal analysis. The selected studies were subsequently exported as NIfTI files to a secure external drive using the Visage application programming interface. Following sequence selection and standardization, HD-BET was applied to extract brain parenchyma from each image, ensuring the removal of identifiable facial features. MRI scans are accompanied by an Excel file containing separate sheets for clinical data and radiologic image acquisition parameters. Each brain metastasis study includes up to four files corresponding to T1W, T1CE, T2W, and/or FLAIR sequences. All imaging data were exported from the Visage AI Accelerator in NIfTI format and processed for brain extraction. File names follow a standardized format, incorporating an anonymous patient identifier, anonymized study date-time, and sequence type, structured as caseID_date-time_sequence.nii.gz to ensure clarity and consistency across the dataset.Abstract
Introduction
Methods
Subject Inclusion and Exclusion Criteria
Data Acquisition
Data Analysis
Anonymization
Usage Notes
Data Access
Version 1: Updated 2025/05/30
June 2025: Updated clinical data sheet to remove duplicates, no significant change to data or imaging.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Radiology Images | MR | NIFTI | Download requires IBM-Aspera-Connect plugin |
1,430 | 33,811 | CC BY 4.0 | — | ||
| Clinical data and Scanner details | Demographic | XLSX | 1,430 | CC BY 4.0 | — |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Chadha, S., Weiss, D., Janas, A., Ramakrishnan, D., Hager, T., Osenberg, K., Willms, K., Zhu, J., Chiang, V., Bakas, S., Maleki, N., Sritharan, D. V., Schoenherr, S., Westerhoff, M., Zawalich, M., Davis, M., Malhotra, A., Bousabarah, K., Deusch, C., Lin, M., Aneja, S., & Aboian, M. S. (2025). Yale longitudinal dataset of brain metastases on MRI with associated clinical data (Yale-Brain-Mets-Longitudinal) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/3YAT-E768 |
Detailed Description
References:
- Achrol, A. S. et al. Brain metastases. Nat Rev Dis Primers 5, 5 (2019).
- Sacks, P. & Rahman, M. Epidemiology of Brain Metastases. Neurosurgery Clinics of North America 31, 481–488 (2020).
- Lamba, N., Wen, P. Y. & Aizer, A. A. Epidemiology of brain metastases and leptomeningeal disease. Neuro Oncol 23, 1447–1456 (2021).
- Brenner, A. W. & Patel, A. J. Review of Current Principles of the Diagnosis and Management of Brain Metastases. Front Oncol 12, 857622 (2022).
- Lin, X. & DeAngelis, L. M. Treatment of Brain Metastases. J Clin Oncol 33, 3475–3484 (2015).
- Vogelbaum, M. A. et al. Treatment for Brain Metastases: ASCO-SNO-ASTRO Guideline. J Clin Oncol 40, 492–516 (2022).
- Le Rhun, E. et al. EANO-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up of patients with brain metastasis from solid tumours. Ann Oncol 32, 1332–1347 (2021).
- Patil, C. G. et al. Whole brain radiation therapy (WBRT) alone versus WBRT and radiosurgery for the treatment of brain metastases. Cochrane Database Syst Rev 9, CD006121 (2017).
- Kraft, J., Zindler, J., Minniti, G., Guckenberger, M. & Andratschke, N. Stereotactic Radiosurgery for Multiple Brain Metastases. Curr Treat Options Neurol 21, 6 (2019).
- Aldawsari, A. M. et al. The role and potential of using quantitative MRI biomarkers for imaging guidance in brain cancer radiotherapy treatment planning: A systematic review. Phys Imaging Radiat Oncol 27, 100476 (2023).
- Kang, T. W. et al. Morphological and functional MRI, MRS, perfusion and diffusion changes after radiosurgery of brain metastasis. Eur J Radiol 72, 370–380 (2009).
- Friedman, D. P., Morales, R. E. & Goldman, H. W. MR imaging findings after stereotactic radiosurgery using the gamma knife. AJR Am J Roentgenol 176, 1589–1595 (2001).
- Lunsford, L. D., Kondziolka, D., Maitz, A. & Flickinger, J. C. Black holes, white dwarfs and supernovas: imaging after radiosurgery. Stereotact Funct Neurosurg 70 Suppl 1, 2–10 (1998).
- Cassinelli Petersen, G. et al. Real-time PACS-integrated longitudinal brain metastasis tracking tool provides comprehensive assessment of treatment response to radiosurgery. Neurooncol Adv 4, vdac116 (2022).
- Aneja, S. & Omuro, A. Imaging biomarkers for brain metastases: more than meets the eye. Neuro Oncol 21, 1493–1494 (2019).
- Aboian, M. et al. Development of a workflow efficient PACS based automated brain tumor segmentation and radiomic feature extraction for clinical implementation (N2.003). Neurology 98, (2022).
- Aboian, M. et al. Clinical implementation of artificial intelligence in neuroradiology with development of a novel workflow-efficient picture archiving and communication system-based automated brain tumor segmentation and radiomic feature extraction. Frontiers in Neuroscience 16, (2022).
- Rudie, J. D. et al. Three-dimensional U-Net Convolutional Neural Network for Detection and Segmentation of Intracranial Metastases. Radiol Artif Intell 3, e200204 (2021).
- Xue, J. et al. Deep learning-based detection and segmentation-assisted management of brain metastases. Neuro Oncol 22, 505–514 (2020).
- Aneja, S., Chang, E. & Omuro, A. Applications of artificial intelligence in neuro-oncology. Current Opinion in Neurology 32, 850 (2019).
- Rudie, J. D., Rauschecker, A. M., Bryan, R. N., Davatzikos, C. & Mohan, S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 290, 607–618 (2019).
Acknowledgements
The authors of this Collection wish to acknowledge:
- Yale Department of Radiology & Biomedical Imaging
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
coming soon |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
