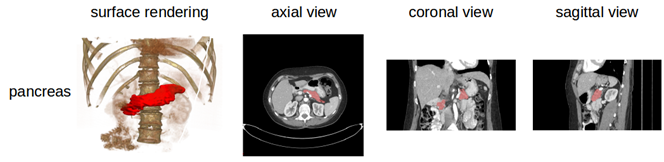
Pancreas-CT | Pancreas-CT
DOI: 10.7937/K9/TCIA.2016.tNB1kqBU | Data Citation Required | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Pancreas | Human | 82 | CT, Segmentations | Healthy Controls (non-cancer) | Image Analyses | Public, Complete | 2020/09/10 |
Summary
The National Institutes of Health Clinical Center performed 82 abdominal contrast enhanced 3D CT scans (~70 seconds after intravenous contrast injection in portal-venous) from 53 male and 27 female subjects. Seventeen of the subjects are healthy kidney donors scanned prior to nephrectomy. The remaining 65 patients were selected by a radiologist from patients who neither had major abdominal pathologies nor pancreatic cancer lesions. Subjects' ages range from 18 to 76 years with a mean age of 46.8 ± 16.7. The CT scans have resolutions of 512x512 pixels with varying pixel sizes and slice thickness between 1.5 − 2.5 mm, acquired on Philips and Siemens MDCT scanners (120 kVp tube voltage). A medical student manually performed slice-by-slice segmentations of the pancreas as ground-truth and these were verified/modified by an experienced radiologist.
Data Access
Version 2: Updated 2020/09/10
Note: Previously posted cases #25 and #70 were found to be from the same scan as case #2, just cropped slightly differently, and were removed from this version of the dataset.
| Title | Data Type | Format | Access Points | Subjects | License | |||
|---|---|---|---|---|---|---|---|---|
| Images | CT | DICOM | Download requires NBIA Data Retriever |
80 | 80 | 80 | 18,942 | CC BY 3.0 |
| Manual Annotations | Segmentations | ZIP and NIFTI | 80 | 80 | CC BY 3.0 |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
- IDC Zenodo community datasets:
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Roth, H., Farag, A., Turkbey, E. B., Lu, L., Liu, J., & Summers, R. M. (2016). Data From Pancreas-CT (Version 2) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.tNB1kqBU |
Detailed Description
Data Example
Note
The DICOM files were created from anonymized volumetric images (Analyze and NifTI) using this from ITK: http://www.itk.org/Doxygen/html/Examples_2IO_2ImageReadDicomSeriesWrite_8cxx-example.html .
Related Publications
Publication Citation |
|
|
Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB, Summers RM. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. N. Navab et al. (Eds.): MICCAI 2015, Part I, LNCS 9349, pp. 556–564, 2015. (arXiv link) https://doi.org/10.1007/978-3-319-24553-9_68 |
Research Community Publications
TCIA maintains a list of publications that leverage TCIA data. If you have a manuscript you’d like to add please contact the TCIA Helpdesk. Below is a list of such publications using this Collection:
- Gibson, E., Giganti, F., Hu, Y., Bonmati, E., Bandula, S., Gurusamy, K., . . . Barratt, D. C. (2017). Towards Image-Guided Pancreas and Biliary Endoscopy: Automatic Multi-organ Segmentation on Abdominal CT with Dense Dilated Networks. Paper presented at the International Conference on Medical Image Computing and Computer-Assisted Intervention.
- Greenspan, H., van Ginneken, B., & Summers, R. M. (2016). Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Transactions on Medical Imaging, 35(5), 1153-1159. doi:10.1109/TMI.2016.2553401
- Shi, H., Lu, L., Yin, M., Zhong, C., & Yang, F. (2023). Joint few-shot registration and segmentation self-training of 3D medical images. Biomedical Signal Processing and Control, 80. doi:https://doi.org/10.1016/j.bspc.2022.104294