Breast-Cancer-Screening-DBT | Breast Cancer Screening - Digital Breast Tomosynthesis
DOI: 10.7937/E4WT-CD02 | Data Citation Required | 7.7k Views | 16 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Breast | Human | 5,060 | MG, Classification, Radiomic Feature, Measurement, Other | Breast Cancer, Non-Cancer | Clinical, Software/Source Code | Public, Complete | 2024/01/18 |
Summary
Breast cancer is among the most common cancers and a common cause of death among women. Over 39 million breast cancer screening exams are performed every year and are among the most common radiological tests. This creates a high need for accurate image interpretation. Machine learning has shown promise in interpretation of medical images. However, limited data for training and validation remains an issue. Here, we share a curated dataset of digital breast tomosynthesis images that includes normal, actionable, biopsy-proven benign, and biopsy-proven cancer cases. The dataset contains four components: (1) DICOM images, (2) a spreadsheet indicating which group each case belongs to (3) annotation boxes, and (4) Image paths for patients/studies/views. A detailed description of this dataset can be found in the following paper; please reference this paper if you use this dataset: M. Buda, A. Saha, R. Walsh, S. Ghate, N. Li, A. Święcicki, J. Y. Lo, M. A. Mazurowski, Detection of masses and architectural distortions in digital breast tomosynthesis: a publicly available dataset of 5,060 patients and a deep learning model. (https://doi.org/10.1001/jamanetworkopen.2021.19100). Additional information and resources related to this dataset can be found here: https://sites.duke.edu/mazurowski/resources/digital-breast-tomosynthesis-database/ A Version 1 of the dataset contains only a subset of all data described in the paper above. More data will be share in subsequent versions. Please visit this discussion forum for any questions related to the data: https://www.reddit.com/r/DukeDBTData/ For some of the images, the laterality stored in the DICOM header and/or image orientation are incorrect. The reference standard "truth" boxes are defined with respect to the corrected image orientation in these instances. Therefore, it is crucial to provide your results for images in the correct image orientation. Python functions for loading image data from a DICOM file into 3D array of pixel values in the correct orientation and for displaying "truth" boxes (if any) are on GitHub. Please see the readme file there for instructions. The DBTex lesion detection challenge tasked participating teams with detecting lesions in the BCS-DBT test set. The challenge had two phases: DBTex1 and DBTex2. Here we provide the BCS-DBT lesion predictions made by all participating teams for both phases, for both the BCS-DBT test and validation sets, as “team_predictions_bothphases.zip”. Please see here under “Output format for the DBTex2 Challenge test set results” for a description of how these results are formatted. Finally, when comparing lesion bounding box predictions to the image data, be sure to load the images correctly according to the above “Required Preprocessing of DBT Images”. If you use these predictions, please reference the DBTex challenge paper: Konz N, Buda M, Gu H, et al. A Competition, Benchmark, Code, and Data for Using Artificial Intelligence to Detect Lesions in Digital Breast Tomosynthesis. JAMA Netw Open. 2023;6(2):e230524. doi:10.1001/jamanetworkopen.2023.0524Required Preprocessing of DBT Images
DBTex Lesion Detection Challenge Predictions
Data Access
Version 5: Updated 2024/01/18
Added spreadsheets:
- Validation set – Spreadsheet indicating which group each case belongs to (see the paper for details on the groups).
- Validation set – Boxes indicating lesion locations.
- Test set – Spreadsheet indicating which group each cases belongs to (see the paper for details on the groups)
- Test set – Boxes indicating lesion locations
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Training set - Images | MG | DICOM | Download requires NBIA Data Retriever |
4,362 | 4,838 | 19,148 | 19,148 | CC BY-NC 4.0 | View |
| Training set - Image paths for patients/studies/views | Other | CSV | 4,362 | 4,838 | CC BY-NC 4.0 | — | |||
| Training set - Spreadsheet indicating which group each case belongs to (see the paper for details on the groups) | Classification | CSV | 4,362 | 4,838 | CC BY-NC 4.0 | — | |||
| Training set - Boxes indicating lesion locations, identical to Phase 1 | Measurement | CSV | 101 | 101 | CC BY-NC 4.0 | — | |||
| Validation set - Images | MG | DICOM | Download requires NBIA Data Retriever |
280 | 312 | 1,163 | 1,163 | CC BY-NC 4.0 | View |
| Validation set - Image paths for patients/studies/views | Other | CSV | 280 | 312 | CC BY-NC 4.0 | — | |||
| Validation set - Spreadsheet indicating which group each case belongs to (see the paper for details on the groups) | Classification | CSV | 280 | 312 | CC BY-NC 4.0 | — | |||
| Validation set - Boxes indicating lesion locations | Radiomic Feature | CSV | 40 | 40 | CC BY-NC 4.0 | — | |||
| Test set - Images | MG | DICOM | 418 | 460 | 1,721 | 1,721 | CC BY-NC 4.0 | View | |
| Test set - Image paths for patients/studies/views | Other | CSV | 418 | 460 | CC BY-NC 4.0 | — | |||
| Test set - Spreadsheet indicating which group each cases belongs to (see the paper for details on the groups) | Classification | CSV | 418 | 460 | CC BY-NC 4.0 | — | |||
| Test set - Boxes indicating lesion locations | Radiomic Feature | CSV | 60 | 60 | CC BY-NC 4.0 | — | |||
| team predictions both phases | Measurement | ZIP and CSV | CC BY-NC 4.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
The following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- Source code: https://github.com/MaciejMazurowski/duke-dbt-data
- https://sites.duke.edu/mazurowski/resources/digital-breast-tomosynthesis-database/
- https://www.aapm.org/GrandChallenge/DBTex
- https://www.aapm.org/GrandChallenge/DBTex2
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Buda, M., Saha, A., Walsh, R., Ghate, S., Li, N., Swiecicki, A., Lo, J. Y., Yang, J., & Mazurowski, M. (2020). Breast Cancer Screening – Digital Breast Tomosynthesis (BCS-DBT) (Version 5) [dataset]. The Cancer Imaging Archive. https://doi.org/10.7937/E4WT-CD02 |
Detailed Description
Dataset Split Details
This dataset was originally used for the DBTex2 challenge (https://www.aapm.org/GrandChallenge/DBTex2/), which contains a total of 22032 breast tomosynthesis scans from 5060 patients from this collection. The dataset was broken down into the following cohorts for usage at the competition, a split that is maintained for this general release:
- Total: 22032 scans
- Training: 19148 scans
- Validation: 1163 scans
- Test: 1721 scans
Version 1, 2 and 3 of the page had a fraction of each of these subsets released, to accompany phase 1 of the aforementioned competition, DBTex.
Training set (with truth):
The training set consists of 19148 cases. This dataset will be representative of the technical properties (equipment, acquisition parameters, file format) and the nature of lesions in the validation and test sets. An associated Excel file in CSV format will include DBT scan identifier and the definition of the bounding box of all lesions.
Validation set (without truth):
The validation set consists of 1163 cases.
Test set (without truth):
The test set consists of 1721 cases.
FREQUENTLY ASKED QUESTIONS
Below are some curated important questions for this dataset along with answers from https://sites.duke.edu/mazurowski/resources/digital-breast-tomosynthesis-database/. For more discussion, visit https://www.reddit.com/r/DukeDBTData/.
Question: Is there a code repository for reading images, drawing bounding boxes, and helper functions related to this database?
Answer: Yes. Please use the following link: https://github.com/MaciejMazurowski/duke-dbt-data
Question: Could you explain the image files/format present for each study?
Answer: One DICOM file or image consists of an entire 3D volume (view). These images are stored in compressed DICOM format.
Question: Which software/tools can I use to read the images?
Answer: You may use a variety of software packages to read the images. We successfully opened the images with the following software: 3D Slicer, ITK-SNAP, Radiant, MicroDICOM, Matlab, and GDCM.
Question: Do I need to know the pre-processing steps, provided in the code repository, for the images?
Answer: It is important to look at the pre-processing steps we provided in the code repository. Please see the Python functions for reading image data from a DICOM file into 3D array of pixel values in the proper orientation and for displaying “truth” boxes (if present). Please also see the readme file there for instructions (https://github.com/MaciejMazurowski/duke-dbt-data). This is crucial as some of the image headers contain incorrect laterality or orientation. For these images, the reference standard “truth” boxes are provided with respect to the corrected image orientation.
Question: Are 4 views available for every study?
Answer: Though 4 views (2 per breast, craniocaudal and mediolateral oblique) are present for most of the studies, some exams have fewer than 4 views.
Question: What kind of encoding is used in the columns of the file ‘BCS-DBT labels-train-v2.csv’?
Answer: The columns “Cancer”, “Benign”, “Actionable”, “Normal” represent one-hot encoded assignment to a category. Details pertaining to these categories can be found in the Section 2.1: https://doi.org/10.1001/jamanetworkopen.2021.19100.
Question: How to interpret the “Slice” column in the data provided in the file ‘BCS-DBT boxes-train-v2.csv’?
Answer: A: The “Slice” column corresponds to the central slice of a biopsied lesion. More details on the image annotation are provided in the paper https://doi.org/10.1001/jamanetworkopen.2021.19100 (section 2.1.2). For evaluation, we assume that lesions span 25% of volume slices in each direction. It is reflected in the evaluation functions available on GitHub: https://github.com/MaciejMazurowski/duke-dbt-data/blob/master/duke_dbt_data.py
Question: How to interpret the columns of ‘BCS-DBT boxes-train-v2.csv’?
Answer:
- PatientID: string – patient identifier
- StudyUID: string – study identifier
- View: string – view name, one of: RLL, LCC, RMLO, LMLO (you might see a numerical suffix after these if multiple images under one view are present)
- Subject: integer – encodes a radiologist who performed annotation
- Slice: integer- the central slice of a biopsied lesion
- X: integer – X coordinate (on the horizontal axis) of the left edge of the predicted bounding box in 0-based indexing (for the left-most column of the image x=0)
- Y: integer – Y coordinate (on the vertical axis) of the top edge of the predicted bounding box in 0-based indexing (for the top-most row of the image y=0)
- Width: integer – predicted bounding box width (along the horizontal axis)
- Height: integer – predicted bounding box height (along the vertical axis)
- Class: string – either benign or cancer
- AD: integer – 1 if architectural distortion is present, else 0
- VolumeSlices: integer – The total number of slices in volume containing the bounding box (used in evaluation function)
Question: Why are the number of bounding boxes much less than the number of training samples?
Answer: The bounding boxes are applicable only to cases with biopsy-proven benign and cancer findings. The training set consists of cases with normal, actionable, benign, and cancer findings. For details on these categories, see Section 2.1: https://doi.org/10.1001/jamanetworkopen.2021.19100.
Question: Why can’t I find the path of a downloaded image in the csv file “Training set – Image paths for patients/studies/views (csv)”?
Answer: At times, you may need to replace “\” with “/” in the path of an image file to find the path in the csv file “Training set – Image paths for patients/studies/views (csv)”.
Question: Why are there both “descriptive_path” and “classic_path” in the .csv file ” Training set – Image paths for patients/studies/views (.csv)”?
Answer: When you download our data using the NBIA Data Retriever, there are two options (“Descriptive Directory Name” and “Classic Directory Name”) for selecting the Directory Type, that correspond to those two paths in the .csv file.
Question: Does the dataset contain microcalcifications?
Answer: It contains microcalcifications. However, they were not annotated and were not the cause for actionability or biopsy.
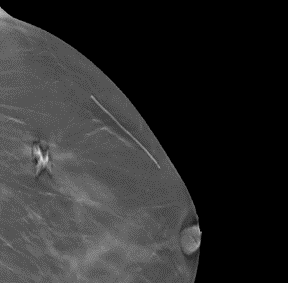
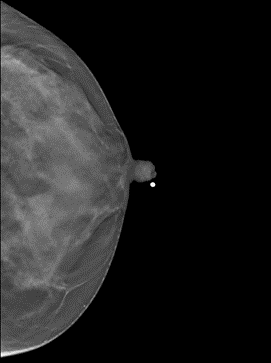
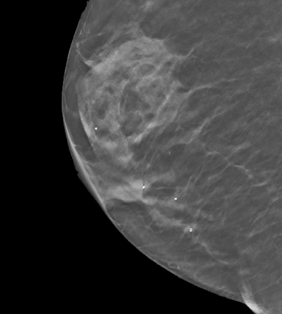
Question: Could you provide some examples of markers in the images?
Answer: Yes. Some examples are listed below. The images or their parts are taken from this collection under the CC BY-NC 4.0 license (https://creativecommons.org/licenses/by-nc/4.0/).
1. Circle for a raised area on the skin such as a mole (image 13345.000000-24122 from this collection)
2. Line for a previous surgery (image 14338.000000-72252 from this collection)
3. Solid pellet for the nipple (image 8764.000000-63613 from this collection)
4. Markers for locations of pain (one or more spots, image 20566.000000-32081 from this collection)
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
Duke University Hospital/Duke University, Durham, NC, USA
We would like to acknowledge all those who contributed to the curation of this dataset
This work was supported by a grant from the NIH: 1 R01 EB021360 (PI: Mazurowski).
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Buda, M., Saha, A., Walsh, R., Ghate, S., Li, N., Swiecicki, A., Lo, J. Y., & Mazurowski, M. A. (2021). A Data Set and Deep Learning Algorithm for the Detection of Masses and Architectural Distortions in Digital Breast Tomosynthesis Images. In JAMA Network Open (Vol. 4, Issue 8, p. e2119100). American Medical Association (AMA). https://doi.org/10.1001/jamanetworkopen.2021.19100, PMC8369362 |
.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 4: Updated 2021/06/08
Added Phase 2 images and spreadsheets. Previous versions (1,2, and 3) are now “Phase 1”.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Training set - Images | MG | DICOM | Download requires NBIA Data Retriever |
4,362 | 4,838 | 19,148 | 19,148 | CC BY-NC 4.0 | — |
| Training set - Image paths for patients/studies/views | CSV | CC BY-NC 4.0 | — | ||||||
| Training set - Spreadsheet indicating which group each cases belongs to (see the paper for details on the groups) | CSV | CC BY-NC 4.0 | — | ||||||
| Training set - Boxes indicating lesion locations, identical to Phase 1 | CSV | CC BY-NC 4.0 | — | ||||||
| Validation set - Images | MG | DICOM | 280 | 312 | 1,163 | 1,163 | CC BY-NC 4.0 | — | |
| Validation set - Image paths for patients/studies/views | CSV | CC BY-NC 4.0 | — | ||||||
| Training set - Spreadsheet indicating which group each case belongs to (see the paper for details on the groups) | CSV | 280 | 312 | CC BY 4.0 | — | ||||
| Validation set - Boxes indicating lesion locations | CSV | 40 | 40 | CC BY 4.0 | — | ||||
| Test set - Images | MG | DICOM | Download requires NBIA Data Retriever |
418 | 460 | 1,721 | 1,721 | CC BY-NC 4.0 | — |
| Test set - Image paths for patients/studies/views | CSV | CC BY-NC 4.0 | — | ||||||
| team predictions both phases | ZIP and CSV | CC BY-NC 4.0 | — |
Version 3: Updated 2021/01/15
Test set – Images added.
Test set – Image paths for patients/studies/views spreadsheet added.
Version 2: Updated 2021/01/04
Validation set – Images added.
Validation set – Image paths for patients/studies/views spreadsheet added.
Volume Slices column added to the Training set – Boxes indicating lesion locations spreadsheet.